Configuring and Running Analysis¶
Sign in to display an analysis window which consists of a navigation bar, a peak list file selection pane and a parameter data configuration pane.
Peak List File Selection¶
[Peak List File]: Drag and drop a file containing peak list data into the text area of [Peak List File]. [Open] is also used to select and upload a peak list file. A maximum of five 200 MB files can be registered. When more files are registered, the oldest ones are removed and replaced by the newer ones. If you are signed in as a guest user, the data will be cleared after you sign out.
MS ASSETs Glycan expect monoisotopic ion’s m/z to analyze and accepts the following files:
“.mgf” or space-delimited text files: These are text files containing peak information separated by line breaks. Each peak’s m/z and intensity values should be listed in that order starting from the beginning of the line, and separated by a space or tab. The data can include a parameter line in “.mgf” format at the beginning of the data, but this will not be used for analysis. The filename extension must be “.mgf” or “.txt”.
“.mzML”: These are “.mzML” files containing peak list information in centroid format with index information. One or more MS 1 peak list in the data is used for the analysis. The filename extension must be “.mzML”.
[History]: Click [History] to select previous uploaded data to analyze. The filenames and registration dates and times of the registered peak lists are displayed in the data selection grid. The newest data is displayed at the top, and will be analyzed by default. Click a different radio button to select another file.
Parameter Data Configuration¶
A parameter data configuration pane consists of a [Parameter Data] and a series of six parameter configuration grids to set analysis conditions. The following sections explain the functions of each configuration.
[Parameter Data]¶
Fill in the name of configuration parameters into the text area of [Parameter Data] and click [Save] to save parameter data into the web application.
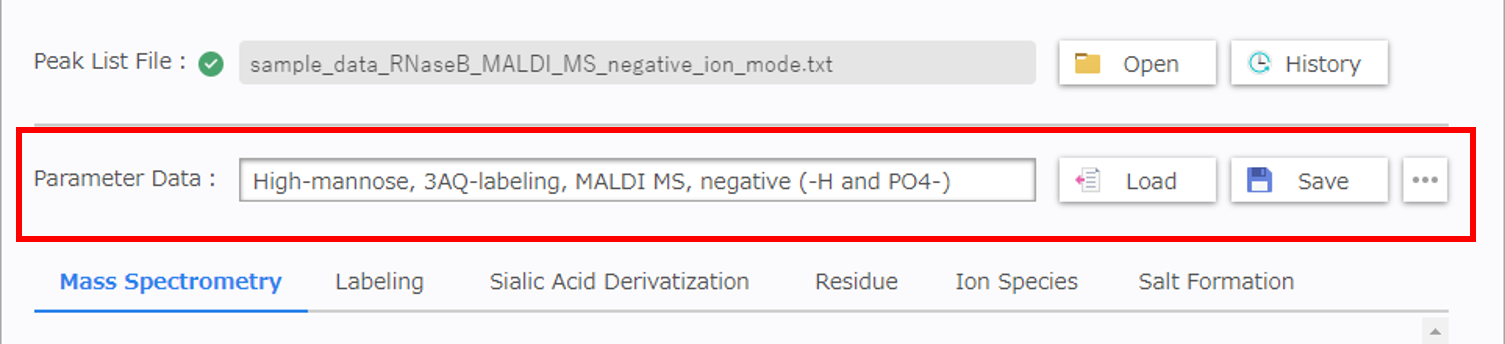
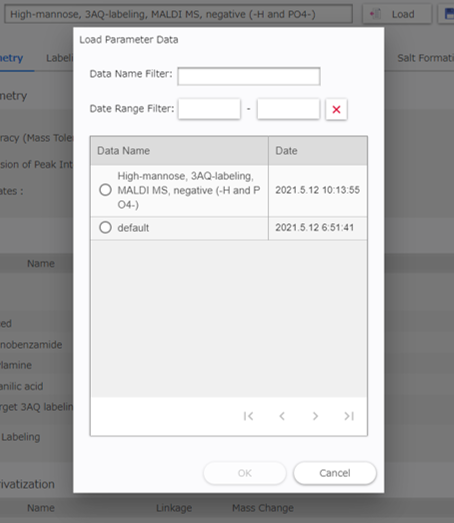
Click [Load] to load a parameter data previously saved.
The saved parameters are displayed in the parameter data grid. You can find previous data by searching for the data name, date and time. Select a data name and click [OK] to load required data.
Click […] - [Save as Default] to use the displayed parameters as default parameters for subsequent analyses.
Note) If you sign in as a guest user, previously saved parameters will be cleared after you sign out.
[Mass Spectrometry Configuration]¶
This configuration is used for ion peak matching.
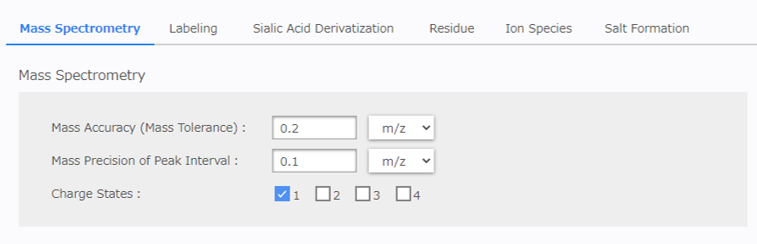
[Mass Accuracy (Mass Tolerance)]: Enter the acceptable m/z tolerance for estimating the composition.
[Mass Precision of Peak Interval]: Enter the acceptable m/z tolerance for matching the mass difference between peaks using the [Siailc Filter]. The value entered for mass accuracy (tolerance) can also be used here.
[Charge State]: Select the charge number. (Multiple selections are allowed.)
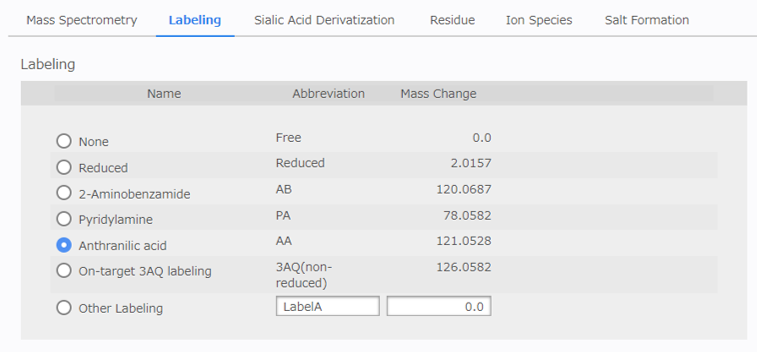
[Labeling Configuration]¶
Select or enter the labeling method for the reducing end of the glycan. Multiple selections are not allowed.
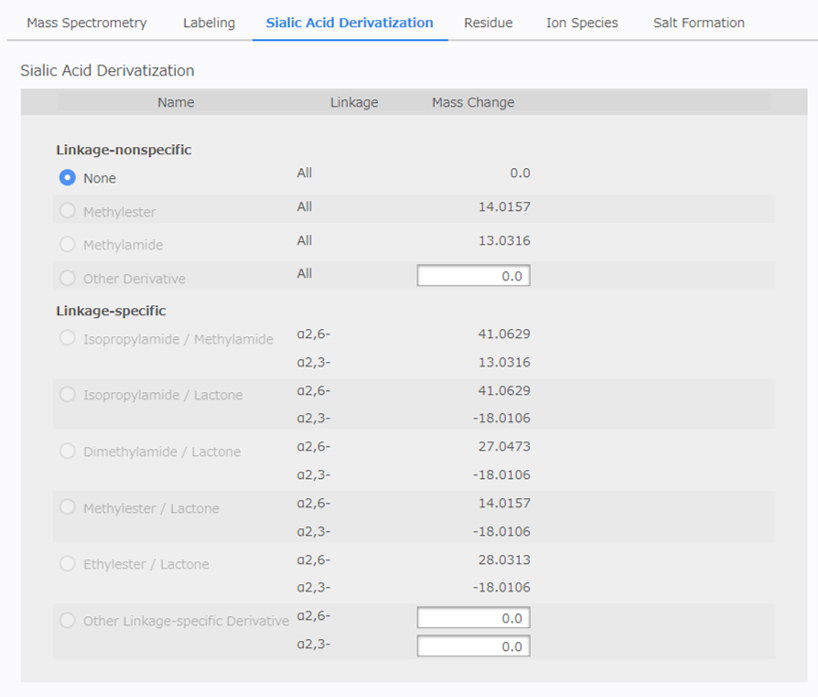
[Sialic Acid Derivatization Configuration]¶
This configuration is enabled just for a user who activated a user option to support sialic acid derivatization. Currently, only SialoCapper TM -ID Kit customers can activate the option.
[Linkage-nonspecific] refers to methods that perform the same derivatization on all sialic acids, and [Linkage-specific] refers to those that perform different derivatization depending on the linkage-type of the sialic acid (α2,3- or α2,6-). Multiple selections are not allowed.
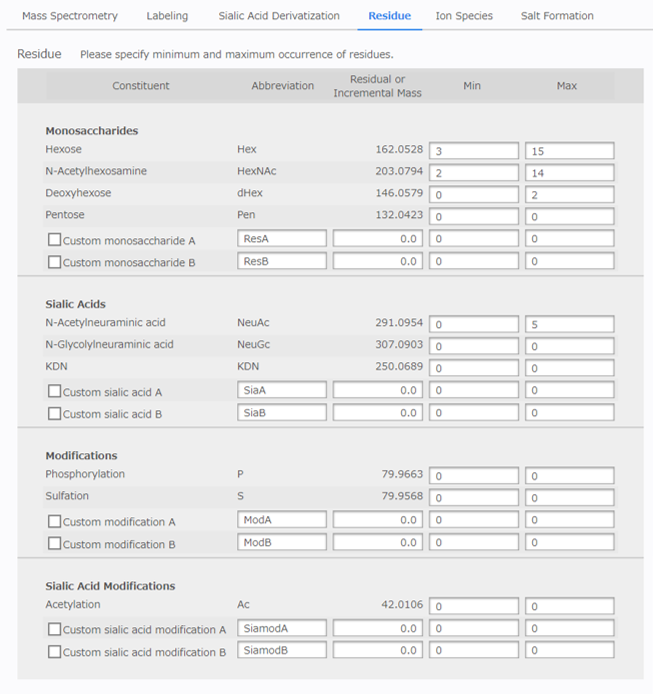
[Residue Configuration]¶
Specify the maximum and minimum occurrence of glycan residues searched for when estimating the composition.
There are two types of sugars listed: [Monosaccharides] and [Sialic Acids]. Sialic acids are subject to mass correction by sialic acid modification. In this application, modifications are handled in the same way as monosaccharides, for the purpose of estimating the composition with consideration to modified glycans such as sulfated glycans.
There are two types of modifications listed: [Modifications], and [Sialic Acid Modifications]. Sialic acid modifications are subject to the mass correction of sialic-acid-containing glycans. It is also possible to specify any residue for each sugar and modification as custom residues. If you want to search for the custom residues, check all box of the residues.
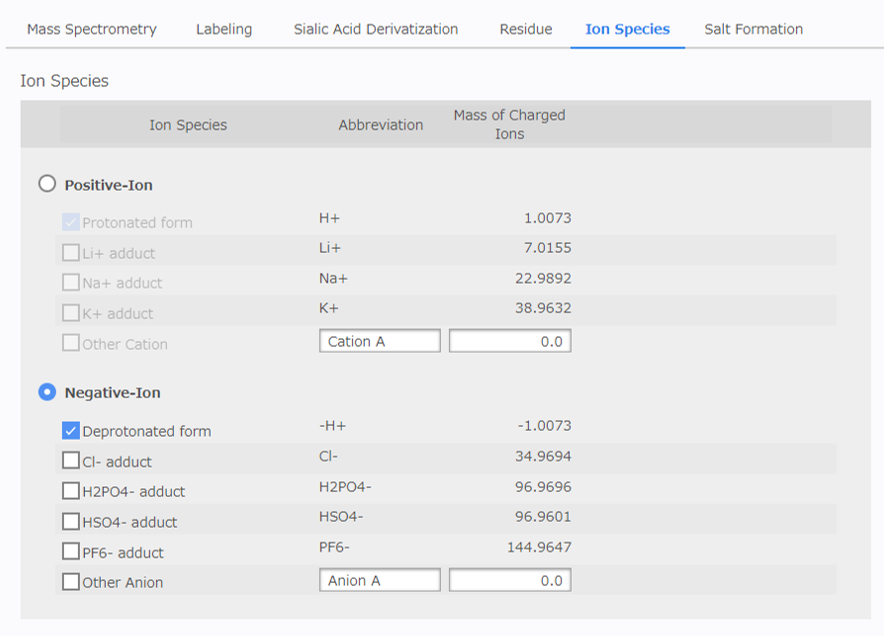
[Ion Species Configuration]¶
Select the ion species to detect. (Multiple selections are allowed.) It is also possible to enter any ion species. Positive and negative ions cannot be selected at the same time.
[Salt Formation Configuration]¶
If the results suggest the possibility of salt formation (salt substitution), it is possible to select candidates 2. If sodium or potassium ions are added but the valence remains the same, select a candidate salt formation in which a proton is replaced by a cation.
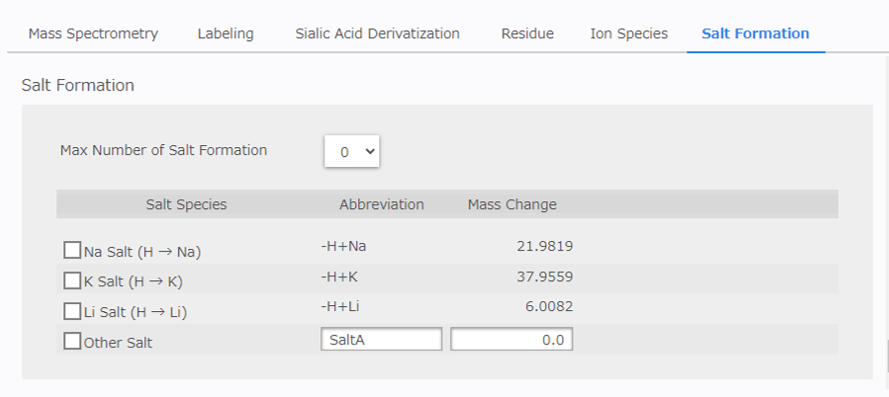
[Max Number of Salt Formation]: Select the maximum acceptable number of salt formations.
[Salt Species] Table: Select the salt formation to consider.
It is also possible to enter other salt formations. In this case, enter the mass difference due to the generation of the salt, not the mass of the salt itself. The mass of the cation is usually -1.0078 because the proton is replaced by the cation.
- 2
If the initial setting for salt formation is too wide, the number of candidates may increase significantly. It is recommended that you start at 0 and increase the value if there is no candidate glycan composition.
Running an Analysis¶
When all conditions have been configured, enter a name in the [Analysis Title] at the navigation bar of the analysis window. The text entered here will be printed at the top of the results window and the previous results dialog. Click [Analyze] and wait for the results to appear.
You need to save or load a parameter data before you perform analysis, if you edit analysis parameters.

